Bacterial glycoprotein antigens for vaccine development
Technology
GlyProVac BEMAP technology
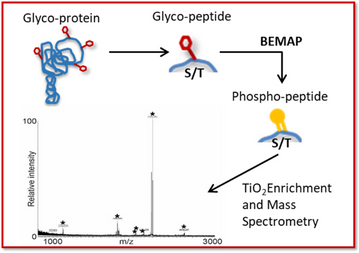
The GlyProVac BEMAP technology is a powerful sample enrichment method based on mass spectrometry, which allows comprehensive identification of any type of O-linked protein glycosylation. BEMAP is an extension of an SDU proprietary method, which was originally developed for enrichment of phosphopeptides. The BEMAP reaction efficiently substitutes O-linked carbohydrate moieties with a 2-aminoethyl phosphonic acid (AEP) group using base-catalysed β-elimination followed by Michael addition of the tag, thus allowing for selective peptide isolation using titanium dioxide.
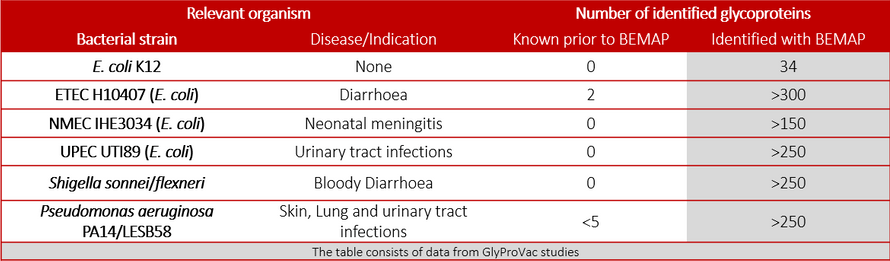
Using BEMAP we have discovered that bacterial O-linked glycosylation is much more extensive than previously thought (see table below) and is especially important to Enterotoxigenic Escherichia coli (ETEC) compared to the commensal E. coli counterpart.
The bacterial monosaccharide building blocks are absent from eukaryotic cells and thus constitute a distinguishing feature of bacteria with important roles in pathogenesis. Most, if not all the protein-based vaccines produced nowadays are produced in the non-pathogen bacterium, thereby disregarding the site-specific O-linked glycosylation.
We believe that O-linked glycoproteins are an emerging reservoir of attractive targets for antimicrobial intervention.
Background
The majority of bacterial O-linked glycoproteins play important roles as mediators of bacterial adhesion to host tissue or as modulators immune responses during infection. Despite their apparent clinical importance, the discovery of novel bacterial glycoproteins implicated in virulence is advancing slowly. This is in part due to inherent challenges associated with glycoproteomics. The analytical tools developed for enrichment of eukaryotic O- and N-linked glycopeptides rely on a limited set of defined physiochemical properties, e.g. glycan hydrophilicity or specific lectin recognition, which are relatively rare in bacteria. The GlyProVac BEMAP technology is the currently only available method to allow comprehensive screening and mapping of bacterial O-linked glycopeptides.